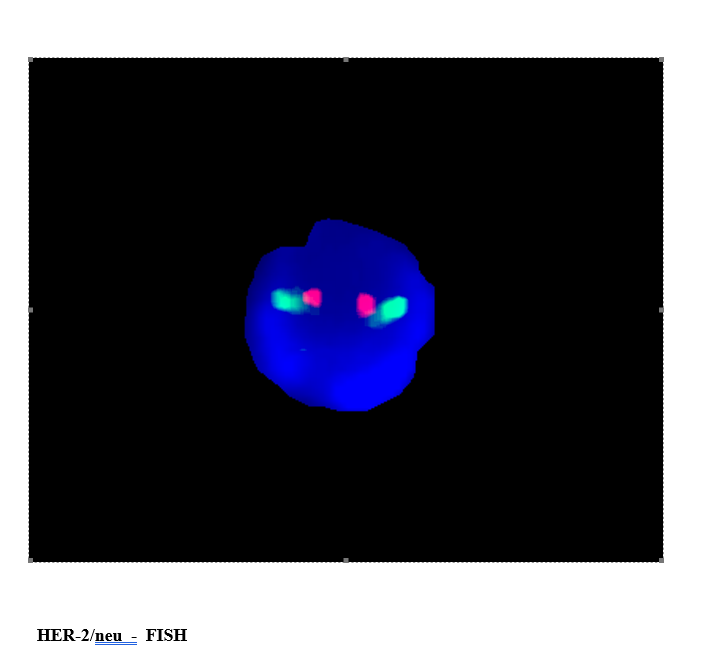
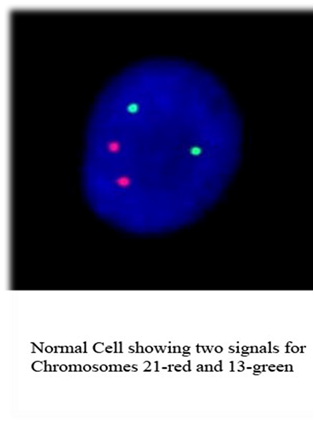
FISH testing techniques
Fluorescence in situ hybridization is a molecular cytogenetic technique where a unique DNA fragment specific for a single chromosome is labeled with a fluorochrome which is visualized under fluorescent microscope using specific filters. A hybridization reaction allows the labeled DNA fragment to bind to its complementary DNA sequences in interphone nuclei/ metaphase chromosomes. Making use of an appropriate filter the presence/absence of chromosomes can be visualized and signals of different colors can be identified. The number of signals indicates the numeric presence of the chromosomes and the signal patterns indicate the presence of structural abnormalities such as translocations involving a specific region.

This technique is very useful inindentifying common aneuplodies (Chromosomes 13, 18, 21, X and Y), micro deletions (Chromosome 22 & 15), specific chromosomal aberrations for oncology conditions [CML panel t(9;22), AML panel {t(8;21), inv 16, MLL, MDS panel (5q, 7q, 20q & CEP 8), BCR/ABL, PML/RARA] and Chimerism study in post BMT analysis.Fixed cell suspensions from venous blood, amniotic fluid or bone marrow samples and formalin fixed paraffin embedded tissue can be used for this technique.
The sample of interest is fixed on the microslide and pretreated to facilitate the penetration of FISH probes. A denaturation step is followed by hybridization with the FISH probes under stringent buffer and temperature conditions. Unbound probes are removed by washing to ensure imaging of specifically stained regions only. Counterstaining of specific cellular structures aids in cell selection. Signal detection is performed on a fluorescence microscope followed by a qualitative or quantitative analysis. This technique is less labor-intensive method for confirming the presence of a DNA segment within an entire genome than other conventional methods like Southern blotting.